Modeling and simulation of biophysical processes
Motivation
Medical images recognition involves consideration of multiple factors responsible for their formation. This knowledge is required because image brightness and contrast depend on both a diagnosed organ state, imaging parameters, as well as interaction between tissue and external signals, such as e.g. RF excitation signal. Moreover, in the case of dynamic imaging sequences, such as perfusion imaging, it is the signal temporal variation which is evaluated. This variation depends on clinically important parameters describing the course of biophysical processes at the tissue, organ and the body scale.
- Research directions
Modeling of biophysical processes aids in computerized image-based diagnosis through automatic determination of their parameters. In addition, simulation of phenomena that take place during image formation helps finding relationships between particular biological and technical parameters on one side, and a measured signal on the other. It then enables prediction of the signal under certain conditions, optimization of an imaging protocol, generation of synthetic data for the need of design and validation of image processing algorithms (segmentation, noise reduction, registration).
- Achievements
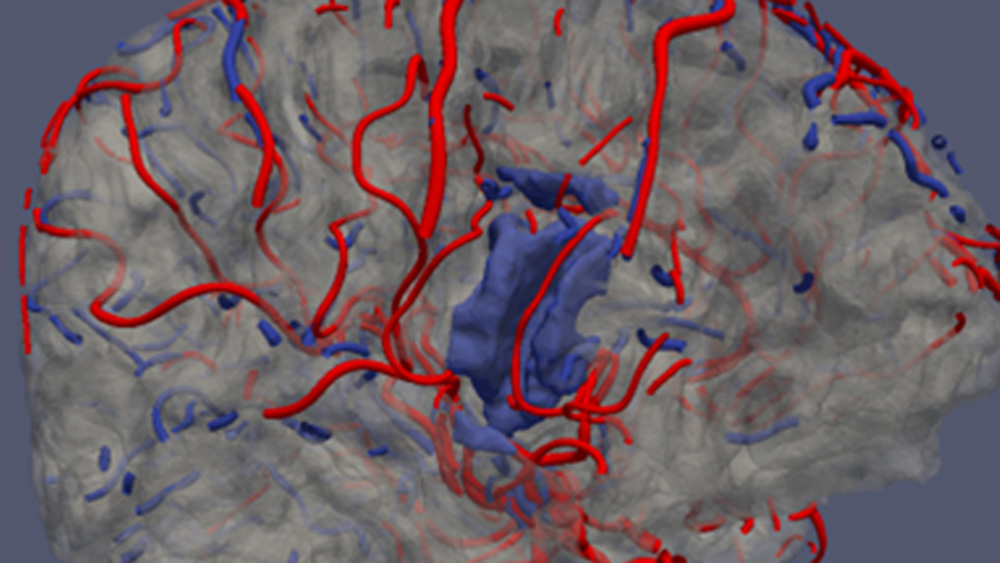
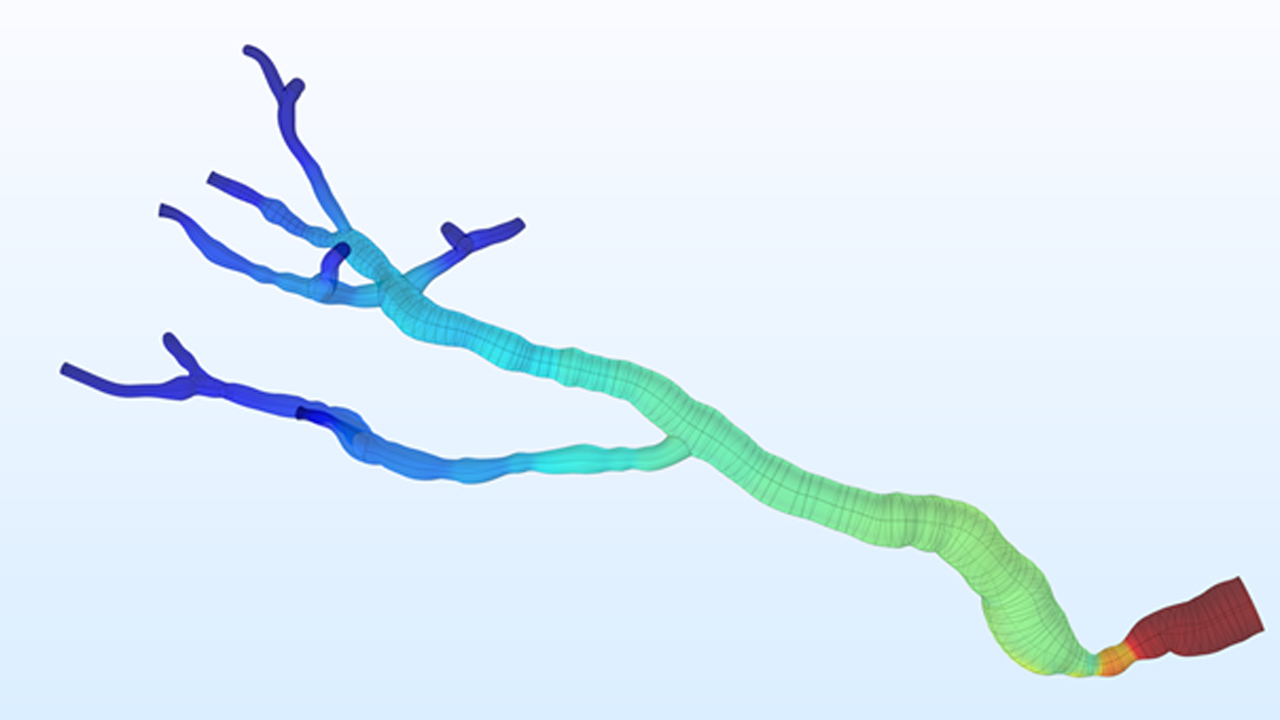
At the Institute of Electronics, we conduct research on modeling the vascular system and simulation of blood flow. These works lead to the design of AngioSim – a computer simulator of magnetic resonance angiography, which enables quantitative validation of results of a segmentation algorithms against a geometrical reference model. We simulated angiographic images not only for realistic models, but also for synthetic vessel trees, constructed according to a specific growth algorithm. Thanks to this achievement, as one of the first research groups, we showed that texture analysis can be applied to describe images of vessels at mesoscale. At the Institute of Electronics, we also work on developing methods for analyzing perfusion-weighted images of brain, kidney, and prostate. For this purpose, we develop software based on pharmacokinetic models, as well as use model-less methods.
- Perspectives
At present, our priority is that the acquired competences in building and applying blood flow and perfusion models can be used in practice. Therefore we intend to continue, in cooperation with radiologists, the development of VesselKnife – a blood vessel segmentation software, and PkModeling – a library for perfusion image analysis.
- Contact persons
- Prof. Andrzej Materka
 , e-mail: This email address is being protected from spambots. You need JavaScript enabled to view it.
, e-mail: This email address is being protected from spambots. You need JavaScript enabled to view it.
- Dr hab. Artur Klepaczko
 , e-mail: This email address is being protected from spambots. You need JavaScript enabled to view it.
, e-mail: This email address is being protected from spambots. You need JavaScript enabled to view it.
- Dr. Marek Kociński
 , e-mail: This email address is being protected from spambots. You need JavaScript enabled to view it.
, e-mail: This email address is being protected from spambots. You need JavaScript enabled to view it.
- Dr. hab. Andrzej Polańczyk

- Prof. Andrzej Materka
- Relevant publications
- Klepaczko, A., Szczypiński, P., Strzelecki, M., Stefańczyk, L. Simulation of phase contrast angiography for renal arterial models (2018) BioMedical Engineering Online, 17 (1), art. no. 41. DOI: 10.1186/s12938-018-0471-y
- Kociński, M., Klepaczko, A., Materka, A., Chekenya, M., Lundervold, A. 3D image texture analysis of simulated and real-world vascular trees (2012) Computer Methods and Programs in Biomedicine, 107 (2), pp. 140-154. DOI: 10.1016/j.cmpb.2011.06.004